Induced mutation
- Induced mutations are induced by known factor- such as-physical (ionizing irradiation, ultraviolet light), chemical and biological mutagens (bacteria and viruses).
Mechanism of induced mutation:
- Induced mutations occurs by at least three different mechanisms. They are-
- By replacing nitrogenous base with base analogs
- By base alteration-altering a base so that it specifically mispair with another base
- By distortion of DNA molecule- damaging a base so that it no longer pair with another base
I. Incorporation of base analogs:
- Some chemical compounds are sufficiently similar to the normal nitrogenous bases of DNA known as base analogs and they can incorporated into DNA in place of normal bases.
- These analogs can base pair with other nitrogenous bases but they induce insertion of incorrect nucleotide during replication causing mutation.
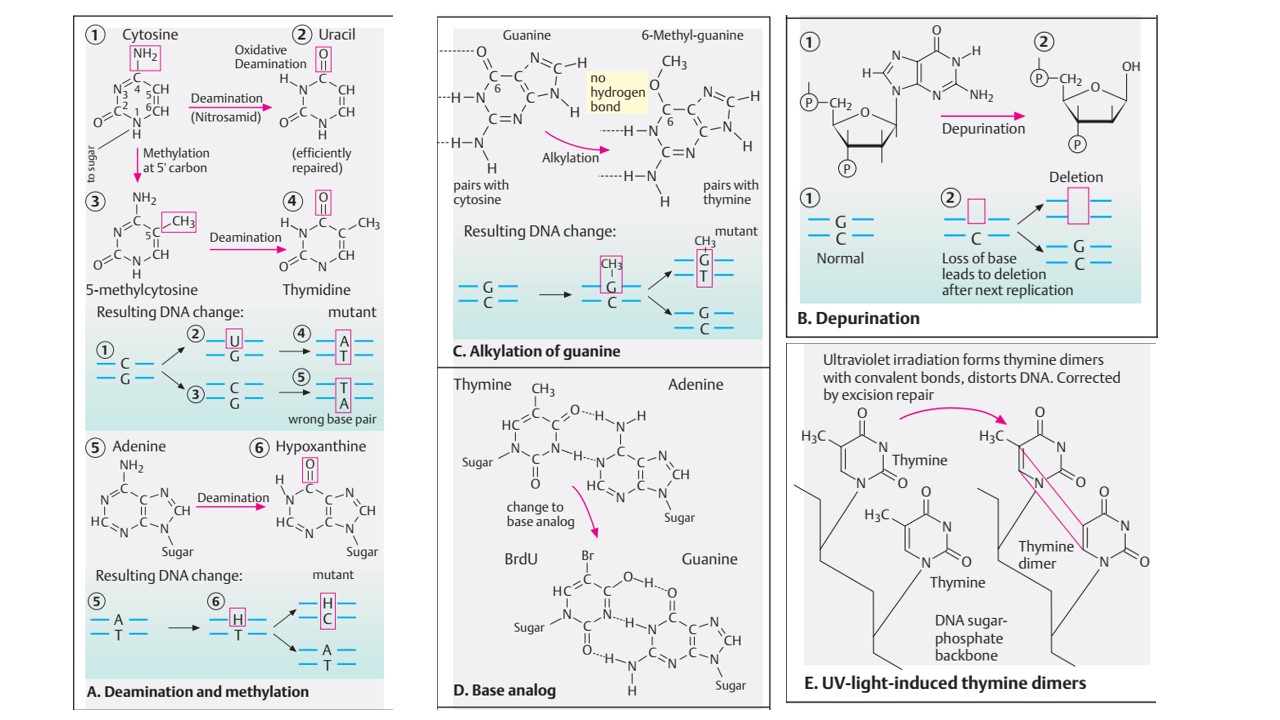
- Uracil is halogenated in the carbon-5 position to give 5-bromouracil, 5-chlorouracil, and 5-iodouracil which can be incorporated into DNA in the place of thymine.
- 5-bromouracil (5-BU) bromine is formed by bromination at the carbon-5 position of uracil. The resulting structure of 5-bromouracil is similar to thymine but thymine has CH3 group at C5 .
- 5-Bromouracil is most effective analog to thymine because size of bromine has same van der Waals radius as the methyl group in thymine.
- 5-bromouracil is highly mutagenic and it pairs with Adenine in normal condition. In 5-BU, the bromine atom is not in a position in which it can hydrogen-bond during base pairing, so the keto form of 5-BU pairs with adenine.
- However the frequency of tautomeric shift of 5-bromouracil is much higher than Thymine. It changes from keto form to either enol form or an ionized from amino form to keto form when protonated. Now, 5-bromouracil form hydrogen bond with Guanine instead of complementary base Adenine which result in base pair transition from T=A to C=G in subsequent replication cycle.
- Another commonly used base is 2-Aminopurine which is analog to adenine and pairs with thymine by two hydrogen bond. After a tautomeric shift due to protonation, it can form pair with cytosine.
- Therefore, 2-aminopurine can induce base pair transition from A=T to G=C in subsequent replication cycle.
II. Base alteration: alkylation, depurination, deamination and hydroxylation
- In this case, the mutagens do not incorporate with the DNA, however they can causes base alteration in a specific way.
- Some of the base alteration are- alkylation, depurination, deamination, hydroxylation etc.
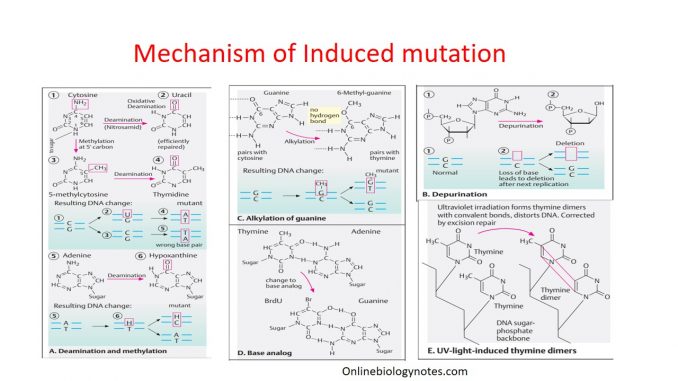
Alkylation:
- Alkylating agents are most widely used mutagens which carry one, two or more alkyl groups in reactive form.
- Alkylating agents transfer their alkyl group (methyl or ethyl) group to nitrogenous bases of DNA and to phosphate group (alkylation).
- The most commonly used alkylating agents are- ethyl-methane sulfonate (EMS), Dimethyl sulphonate (DMS), Diethyl sulphonate (DES) etc , nitrosoguanidine (NG)
- Mechanism of alkylating agents:
- Transfer alkyl group to phosphate group of DNA and produces unstable phosphate trimester which hydrolyses to give alkyl group. But some alkyl group may remain attached to phosphate which interfere with DNA replication cycle as well as cause breakage of sugar phosphate back bone.
- Transfer alkyl group to 6-oxygen and 7-nitrogen of DNA bases. Among these alkylation of nitrogenous base, 7-ethylguanine is most common derivative. 7-ethylguanine is base analog to cytosine and can form base pair with thymine which ultimately cause base transition from G=C to AT.
- Some difunctional or polyfunctional alkylating agents can crosslink with DNA strand and interfere DNA replication leading to chromosome breakage.
Depurination:
- Alkylation of purine base (Adenine and Guanine) give rise to unstable quaternary nitrogenous base which interfere with glycosidic bond between nitrogenous base and deoxyribose sugar.
- The loss of purine bases from DNA is termed as Depurination.
- Once the base is depurinated, it cannot specify complimentary base to the original purine during replication which result in incorporation of wrong base.
- Therefore, this causes base pair substitution.
Deamination:
- Some mutagens react with nitrogenous base containing amino group and remove it with another functional group.
- Nitrous acid (HNO2) a chemical which reacts with amino group containing nitrogenous bases (Adenine, cytosine and guanine) and replace it.
- Deamination of adenine by nitrous acid yields hypoxanthine (H) which can base pair with cytosine. This deamination at the position of adenine result in base transition from A:T to G:C in successive replication.
- Similarly, deamination of cytosine by HNO2 results in uracil. Now uracil can base pair with adenine. Therefore, change of base from cytosine to uracil results in base transition from G:C to A:U and then A:T in successive replication cycle.
- Other example of deamination: conversion of 5-methylcytosine to thymine.
Hydroxylation:
- The mutagen hydroxylamine (NH2OH) reacts with amino group of cytosine causing hydroxylation to yield hydroxylcytosine.
- Now the hydroxylcytosine can form base pair with adenine instead of guanine. This results in base pair transition.
III. Distortion of DNA molecule:
- Some of the mutagens directly reacts with DNA molecules causing distortion and modify the structure.
i. Intercalating agents:
- Certain florescent acridine dyes such as acridine orange, proflavin causes DNA mutation by insertion or deletion of nitrogenous bases.
- Acridine dyes are planar (flat) molecule that mimic nitrogenous bases and at low concentration it can inserts or intercalates between subsequent nitrogenous bases in DNA molecule.
- Insertion of the agent stretches the distance between adjacent base pair by 0.68nm which is twice the normal distance. This distortion result in single nucleotide deletion or insertion at this position during recombination.
- Intercalating agents result in insertion of bases:
- Intercalation of acridine dye between two nitrogenous bases in template strand result in stretch of DNA molecule.
- During DNA replication, a new base can be inserted in the newly synthesized strand opposite of acridine molecule.
- Now during replication of newly synthesized strand, a complimentary base in added opposite to newly added base.
- Intercalating agents result in deletion of bases:
- Insertion of acridine molecule in DNA strand may block the base in the template strand and does not allow to base pair.
- During replication, one base is deficient in newly synthesized strand.
ii. DNA damage: by radiation
- Various kinds of radiations can induce mutation. Mutagenic radiations are two types- non-ionizing radiation (UV rays) and ionizing radiation (X-rays, gamma rays):
a. Mutation caused by UV rays: Pyrimidine dimer formation
- UV rays have a wavelength of 10-390nm.
- When UV rays falls on the genetic materials, it is absorbed and due to increase in energy level electrons are excited.
- The most effective wavelength of UV rays for inducing mutation is 260nm.
- UV rays are non-ionizing radiation and have a penetration power lower than X-rays.
- Exposure of DNA to UV rays causes nitrogenous bases to become highly reactively unstable free radicals.
- One of the consequences of UV rays exposure is photochemical fusion of two pyrimidine that are adjacent to each other on a same polynucleotide chain.
- Pyrimidine dimer formation is the primary effect of UV rays on DNA molecule.
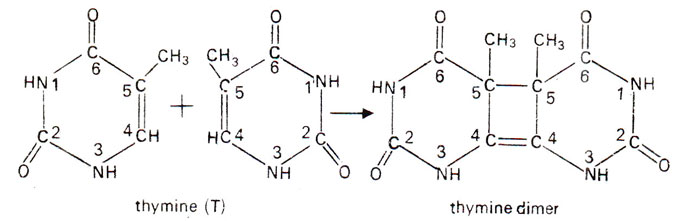
- In case of two thymine, the fusion is called thymine dimer. UV rays causes adjacent thymine on the same DNA strand to bond together by covalent linkage between carbon 5 and 6 of adjacent thymine molecule forming cyclobutane ring.
- In case of thymine adjacent to a cytosine, the resulting fusion is a thymine-cytosine dimer in which the thymine is linked via its carbon atom 6 to the carbon atom 4 of cytosine.
- This dimer structure cannot fit into DNA helix and cause distortion of DNA molecule which result in failure of DNA replication and sometime leads to lethal effect.
- Pyrimidine dimer can also formed by cross linking of pyrimidine bases of adjacent DNA strand.
- There are three types of thymine dimers
- Thymine-thymine dimer= 50%
- Thymine-cytosine dimer= 40%
- Cytosine-cytosine dimer=10%
b. Mutation caused by Ionizing radiation (IR):
- The effects of different ionizing radiation are qualitatively the same.
- In direct effect, IR breaks the phosphate ester bond in DNA. The breakage may take place at one or more points. As a result of breakage, segment of DNA either get lost or rearranged during repair. Sometimes the effect is fatal.
- In indirect effect, the ionizing radiation ionize water producing free radicals which is extremely reactive.
- The free radicals react with DNA molecule to alter its structure.
