Antisense technology:
- Antisense technology is a recent approach to specific modification or inhibition of gene expression in vitro or in vivo.
- It is a tool to study gene function and utilize it to manipulate the gene expression within cells to treat an endless number of diseases.
- The antisense approach utilizes antisense agents to alter the expression of viral genome inside the host cell or regulate the expression of specific genes that causes that particular disease.
- Sense strand or sequence: it is the coding strand within double-stranded DNA that carries the translatable code in the 5′ to 3′ direction. It is complementary to the template strand. The sense strand have sequences similar to that of mRNA.
- Antisense strand: it is the template strand of ds DNA, from which mRNA is transcribed. Thus, the antisense strand is complementary to mRNA.
- In simple term ‘sense’ refers to original sequence of DNA or RNA molecule and ‘antisense’ refers to Complementary copy.
- The antisense strand base pairs with its complementary mRNA strands (sense RNA) and thus prevents it from being translated into a protein.
- Therefore in antisense technology, the complementary nucleic acid sequence (antisense agents) is utilized to silence gene expression. The binding, or hybridization, of antisense nucleic acid sequences to a specific mRNA target will inhibit normal gene expression (ie. either interrupt transcription or translation) resulting in flow of message from DNA to protein.
- There are several mechanisms to interrupt the gene expression by antisense technology. Sometime the gene expression is completely inhibited known as knock-out and other time it is partially interrupted known as knock-down.
- Some of the antisense agents are:
- Antisense oligonucleotide: like oligodeoxyribonucleotides (ODN) having less than 30 mucleotides or longer antisense RNA (a RNA) sequences
- First generation
- Second generation
- Third generation
- Ribozymes
- RNA interference (RNAi)
- Antisense oligonucleotide: like oligodeoxyribonucleotides (ODN) having less than 30 mucleotides or longer antisense RNA (a RNA) sequences
1. Antisense oligonucleotide:
- Zamecnik and Stephenson first demonstrated the antisense effect of synthetic oligonucleotide
- Zamecnik and Stephenson identified a repeated sequence of 21 nucleotides that was crucial to viral integration with the help of nucleotide sequences from the 5’ and 3’ ends of the 35S RNA of Rous sarcoma virus (RSV).
- They synthesized a 13-mer oligonucleotide, d(AATGGTAAAATGG), complement to the portion of this viral sequence.
- Viral production got inhibited when synthetic oligonucleotide was introduced into cultured fibroblast cells. Thus, they concluded that oligonucleotide was inhibiting viral integration by hybridizing to the crucial sequences and blocking them. They introduced the term ‘hybridon’ to describe such oligonucleotides.
- At the same time, Tennant et al and Miller et al reported similar effects for synthetic oligonucleotides in other systems.
- Criteria
for successful oligonucleotide
- Specific target recognition by Watson-Crick pairing
- Good structural Mimicry
- Activation of RNaseH
- Enhanced cellular uptake
- Enhanced resistance to various nucleases: Synthetic oligonucleotides are foreign to the cells into which they are introduced and thus becomes prey for endogenous nucleases.
- Synthetic oligonucleotides were protected from endogenous nuclease when they attained the persistence level in cell.
- There are three possible sites on a nucleotide where protective modifications could be introduced.
Antisense Oligonucleotide modification: first, second and third generation
- The three possible sites for oligonucleotide modification are- at the position of Nitrogenous Base, Ribose sugar (2’ OH group) and the Phosphate backbone.
- The main purpose of modification is to protect the antisense nucleotide from nuclease degradation when introduced inside cells. And at the same time it should be considered that the modification do not alter the inhibit hybridization ability of the antisense nucleotide.
First generation modification:
- The first generation antisense-motivated nucleotide modification is made by Eckstein and colleagues in the late 1960s by replacing one of the oxygen atom (non-bridging oxygen) of the phosphate backbone with a sulfur atom.
- This modified antisense agent is known as phosphorothioate.
- Phosphorothioate oligonucleotide is more nuclease resistant than the original oligonucleotide. The nuclease resistance was measured by an increased half-life for a phosphorothioated oligonucleotide upto ten hours in human serum as compared to that of one hour of an unmodified oligonucleotide having the same sequence.
- However, the phosphorothionated nucleotide displayed slight reduced hybridization ability and also a tendency to bind un-specifically to certain proteins in cell. High concentration of phosphrothionated nucleotide thus result in cytotoxicity.
- Matsukura and colleagues demonstrated that phosphorothioated oligonucleotides were effective hybridons against the HIV replication in the cultured cells.
- The first FDA-approved antisense drug is Vitravene from ISIS (Carlsbad, CA, USA.)
Second generation modification:
- The second generation modification focused on non-specific bind with certain proteins and cytotoxic effect of phosphorothioated nucleotides.
- In this modification, the antisense oligonucleotide undergoes alkyl modifications at the carbon no 2 (C2 position) of the ribose sugar.
- The two most important of these modifications are 2’-O-methyl and 2’-O-methoxy-ethyl at the C2 position.
- After alkylation at the C2 position of ribose sugar, the antisense oligonucleotides become resistant to nuclease degradation and shows low cytotoxicity effect.
- However, the antisense oligonucleotide with 2’-O-alkyl modification are unavailable for RNase H cleavage after hybridization with sense RNA.
- Since RNase H cleavage is the most desirable mechanism for antisense effect, A hybrid oligonucleotide is constructed containing both the desirable characteristics of nuclease resistance and RNase cleavage and it is known as gapmer antisense oligonucleotide.
- Gapmer
antisense oligonucleotide:
- This hybrid antisense oligonucleotide contains a central block of deoxynucleotides sufficient to induce RNase H cleavage flanked by blocks of 2’-O-methyl modified nuclease resistance ribonucleotide.
Third generation modification:
- In this modification a.
- Antisense oligonucleotide forms either DNA: DNA homo-duplex or DNA: RNA hetero-duplex depending upon the nature of oligonucleotide.
- The unmodified oligo-deoxynucleotides form such desired DNA: DNA or DNA:RNA duplexes.
- However variety of nucleic acid analogs have been developed having high affinity with target DNA or RNA and as a modification these analogs are utilized for antisense oligonucleotide construction.
- Some of these nucleic acid analogs are peptide nucleic acids (PNAs), 2’-fluoro N3-P5’-phosphoramidites, 1’, 5’- anhydrohexitol nucleic acids (HNAs) and locked nucleic acids.
- Among these analogs locked nucleic acid (LNA) is very desirable as shows promising effect. The LNA is composed of nucleotides that is locked into a single conformation through a 2’-0’, 4’-C methylene linkage in 1,2:5,6-di-O-isopropylene-α-allofuranose. LNA has increased the thermodynamic stability and enhanced nucleic acid recognition.
2. Ribozymes:
- Ribozymes are known as catalytic RNA, first described by Tom Cech (1982) from ribosomal RNA precursor from Tetrahymena thermophilia.
- Ribozymes acts as an enzymes to processes RNA precursors. And catalyze the modification or alteration of RNA or even DNA.
3. RNA Interference (RNAi):
- RNA interference (RNAi) was first described by Fire and colleagues in Caenorhabditis elegans.
- Several types of very short RNAs repress or silence the expression of genes and such silencing is known as RNA interference.
- RNA interference manifest in different ways; some time by inhibiting translation of mRNA and in other case by destruction of mRNA or silencing of promoter.
Antisense mediated gene silencing
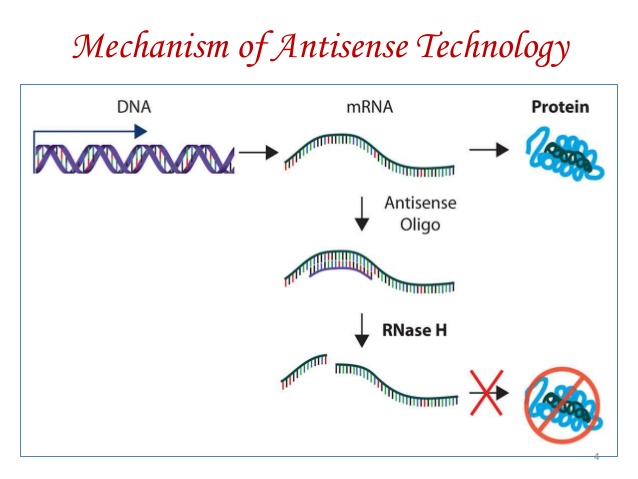
- The basic concept in gene silencing utilizing antisense technology is that an antisense oligonucleotide (a short DNA or RNA) is synthesized and introduced into a cell. Since the antisense oligonucleotide is complementary to the targeted mRNA, it will bind forming RNA dimers in the cytoplasm and halts protein synthesis. This occurs because the mRNA no longer has access to the ribosome and cytoplasm. In the same time, it is degraded by nucleases.
- Therefore, the introduction of antisense oligonucleotide results in silencing the expression of gene.
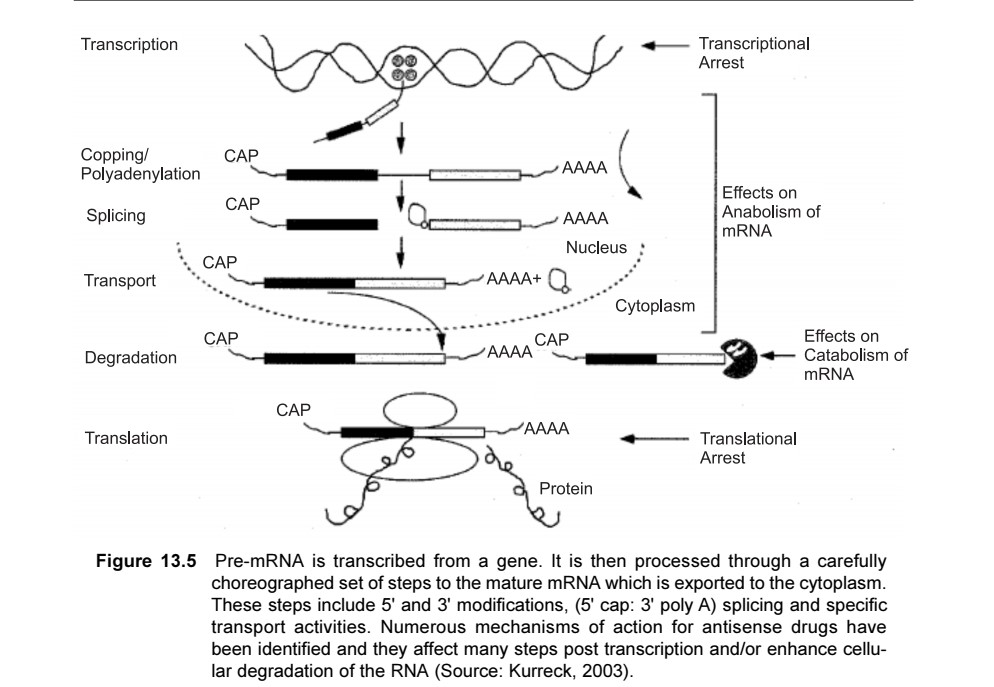
- The events of gene expression is the flow of information from DNA into proteins through transcription (post transcriptional modification) and translation (post-translational modification).
- In the first step DNA is transcribed into pre-mRNA
- Pre-mRNA undergoes post transcriptional modification including- 5’ capping, removal of intron (intron excision) and poly-adenylation to form mature functional mRNA.
- Then mRNA is transported to ribosome for translation.
- For silencing gene expression, the antisense oligonucleotide acts on each step of gene expression to achieve antisense knock-down or knock-out of the targeted gene.
- Gene silencing by antisense mechanism includes-Blocking RNA splicing, accelerating degradation of the RNAmolecule, and preventing introns from being spliced out of the mRNA, impeding the exportation of mRNA into the cytoplasm, hindering translation, and the triplex formation in DNA.
Silencing transcription:
- In this step, antisense oligonucleotide turn off the gene expression by binding either of the three region- minor groove binding, Strand displaying by nucleic acid analogs, and major groove binding , triplex forming oligonucleotides.
- Pyrrole-imidazole are minor groove binding antisense polypeptides that binds with specific sequence in minor groove.
- Nucleic acid analogs containing antisense oligonucleotide binds with complementary strand of DNA helix, displacing the other strand. This is because the affinity and stability of nucleic acid analog with DNA is more than DNA: DNA duplex.
- Some antisense oligonucleotide are triplex forming and they create stable triplex DNA helix. Triplex forming oligonucleotides binds to duplex through Hoogsteen hydrogen binding: T-A:T and C-G:C triplets.
Silencing post-transcription:
- In this step, antisense oligonucleotides inhibit post-transcriptional modification or RNA splicing.
- Once the mRNA is transcribed from DNA, it undergoes several modification including 5’ capping, polyA tail and intron removal. After post transcriptional modification, mRNA is transported out from nucleus to cytoplasm. Once the mRNA is hybridized with antisense oligonucleotide, the double stranded RNA duplex cannot be transported out to cytoplasm.
- Removal of intron is essential for RNA splicing because intron are non-coding sequences. In this process, the oligonucleotide-based antisense agent is used which binds with specific sequence of pre-mRNA preventing intron-excision.
Silencing translation:
- In this step, antisense oligonucleotide inhibit translation by complement pairing with targeted mRNA.
- Binding of antisense oligonucleotide with mRNA results in duplex formation which interferes with the transcription apparatus and prevents formation of the ribosomal complex. Thus translation is halted.
- When an antisense strand binds to a mRNA strand, it form a double helix structure which is recognized as faulty and degraded by RNase H preventing the production of undesired protein.
- RNase H degradation mechanism is most used mechanism in gene silencing. RNase H is an endogenous enzyme which cleaves the RNA moiety of an RNA: DNA duplex.
- RNase is present in both nucleus and cytoplasm. It cannot degrade single stranded mRNA but can degrade dsRNA duplex.