Messenger RNA (mRNA) and its region
- Messenger RNA operates as the template for protein synthesis.
- Messenger RNA encodes genetic information from DNA as a transcript and translates the information to a ribosome and helps assemble amino acids in their proper order.
- mRNA is directly transcribed from DNA, whereas in case of eukaryotes, a pre-mRNA (also referred to as the primary transcript) is first transcribed from DNA and then processed to yield mature mRNA.
- Three main regions occur in both prokaryotic and eukaryotic mRNAs.
- 5’ UTR:
- The 5′ untranslated region (5′ UTR; also called the leader) is a nucleotide sequence at the 5′ end of the mRNA that does not encode any of amino acids.
- In bacterial mRNA, this region consists of the consensus sequence termed as the Shine-Dalgarno sequence. During translation, Shine-Dalgarno sequence serves as a ribosome binding site. This sequence is found approximately seven nucleotides upstream of the first codon that is translated into the amino acid, also termed as start codon.
- In its 5′ untranslated region, eukaryotic mRNA has no equivalent consensus sequence.
- Ribosomes bind to a modified 5′ end of mRNA in eukaryotic cells.
- Protein coding region:
- The next section of mRNA is the protein-coding region, containing the codons that describe the protein’s amino acid sequence.
- The protein-coding region starts with a start codon and terminates with a stop codon.
- 3’ UTR:
- The 3′ untranslated region (3′ UTR; also referred to as a trailer), a nucleotide sequence at the3′ end of the mRNA, is the last mRNA region and not translated into protein.
- The3′ UTR affects mRNA stability and the translation of the protein-coding sequence of the mRNA.
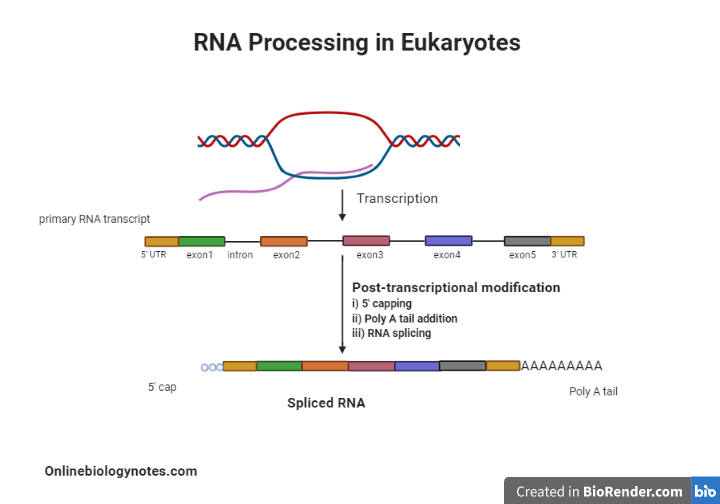
Post-transcriptional modification in Eukaryotes
- Transcription and translation take place concurrently in bacterial cells; when the 3′ end of an mRNA is undergoing transcription, ribosomes bind near the 5′ end to the Shine-Dalgarno sequence and begin translation.
- Since transcription and translation are coupled, before protein synthesis, bacterial mRNA has little opportunity to be changed.
- In contrast, in eukaryotic cells, transcription and translation are segregated both in time and space.
- In the nucleus, transcription takes place while translation takes place in the cytoplasm; this separation offers a chance to modify eukaryotic RNA before translating it.
- Indeed, after transcription, eukaryotic mRNA is altered extensively.
- Changes are made to the RNA molecule’s 5′ end, the 3′ end, and protein coding portion.
- Following are the examples of Post-transcriptional modification:
1. The 5 ‘Cap Addition:
- One type of eukaryotic pre-mRNA modification is the addition of a structure called a 5 ‘cap at its 5’ end.
- At the 5’ end of the mRNA, the cap consists of an additional nucleotide and methyl groups (CH3) at the base of one or more nucleotides at the 5′ end of the newly inserted nucleotide and the 2′-OH group of sugar.
- After transcription initiation, the insertion of the cap takes place quickly.
- It is possible to represent the 5′ end of pre-mRNA as 5′-pppNpNpN, in which a ribonucleotide is represented by the letter ‘N‘ and a phosphate by ‘p‘.
- One of these phosphate groups is removed shortly after the start of transcription and a guanine nucleotide is added.
- A special 5′-5′ bond connects this guanine nucleotide to the pre mRNA, which is somewhat different from the normal 5′-3′ phosphodiester bond that binds all the other RNA nucleotides.
- To the 5′ end, one or more methyl groups are added.
- The first of these methyl groups is attached to the position 7 of the base of the terminal guanine nucleotide making the base 7-methyl guanine.
- Next, in the second and third nucleotides, a methyl group may be attached to the 2′ position of the sugar.
- Additional methyl groups can rarely be attached to the bases of the second and third nucleotides of pre-mRNA.
2. The Poly A tail addition:
- The addition of 50 to 250 or more adenine nucleotides at the3′ end, forming a poly(A) tail, is a second kind of modification to eukaryotic mRNA.
- These nucleotides are not encoded in the DNA, but are inserted in a process called polyadenylation following transcription.
- Many RNA polymerase II transcribed eukaryotic genes are transcribed well past the end of the coding sequence; much of the extra material is then cleaved at the3′ end and the poly(A) tail is inserted.
- Sequences both upstream and downstream of the cleavage site are necessary for processing the3′ end of pre-mRNA.
- Generally, downstream of the cleavage site is a sequence rich in uracil nucleotides (or in guanine and uracil nucleotides).
- On many mRNAs, the poly(A) tail confers stability, increasing the time during which the mRNA remains intact and available for translation before cellular enzymes degrade it.
- The poly(A) tail also enhances the ribosome’s attachment to the mRNA.
3. RNA splicing:
- The removal of introns by RNA splicing is the other major type of eukaryotic pre-mRNA modification.
- Before the RNA moves to the cytoplasm, this modification takes place in the nucleus.
- The presence of three sequences in the intron is required for splicing.
- One end of the intron is referred to as the5′ splice site, and the other end is the 3′ splice site; short consensus sequences exist on these splice sites.
- Most introns begin with GU in pre-mRNAs and end with AG.
- The third sequence that is necessary for splicing is present at the branch point, which is an adenine nucleotide that is situated 18-40 nucleotides upstream of the 3′ splice site.
- Splicing occurs within a large structure called the spliceosome, which is one of the largest and most complex of all molecular complexes.
- Five RNA molecules (U1, U2, U4, U5, and U6) and almost 300 proteins form the spliceosome.
- Small nuclear RNAs (snRNAs) ranging in length from 107 to 210 nucleotides are the RNA components; these snRNAs are associated with proteins to form small particles of ribonucleoprotein.
Process of RNA splicing:
- An intron is between an upstream exon (exon1) and a downstream exon (exon 2) before splicing takes place.
- In two distinct stages, pre-mRNA is spliced.
- The pre-mRNA is cut at the 5 ‘splice site in the first stage of splicing.
- This cut frees exon 1 from the intron, and the intron’s 5′ end connects to the branch point; that is, the intron folds back on itself, creating a structure called a lariat.
- In this reaction, via a trans-esterification reaction, the guanine nucleotide in the consensus sequence at the 5′ splice site binds with the adenine nucleotide at the branch stage.
- To the cytoplasm, where it is translated, the mature mRNA consisting of the exons spliced together is exported.
- A cut is made at the3′ splice site in the second step of RNA splicing and, simultaneously, the3′ end of exon 1 is covalently connected (spliced) to the5′ end of exon 2.
- It releases the intron as a lariat.
- When the bond splits at the branch stage, the intron becomes linear and is then quickly degraded by nuclear enzymes.
- To the cytoplasm, where it is translated, the mature mRNA consisting of the exons spliced together is exported.
Alternative processing pathways for RNA splicing:
- In order to generate alternative forms of mRNA, a single pre-mRNA is processed in various ways, resulting in the development of various proteins from the same DNA sequence.
- Alternative splicing, in which the same pre-mRNA can be spliced in more than one way to generate multiple mRNAs that are translated into different amino acid sequences and thus different proteins, is one form of alternative processing.
- Another method of alternative processing involves the use of several 3′ cleavage sites, where the pre-mRNA comprises two or more potential cleavage and polyadenylation sites.
- In the same pre-mRNA transcript, both alternative splicing and multiple 3′ cleavage sites can exist.
- In multicellular eukaryotes, alternative processing of pre-mRNAs is common.
- Researchers predict, that more than 90% of all human genes undergo alternate splicing.
- The type of splicing also varies between human tissues; compared to other tissues, the human brain and liver tissues have more alternatively spliced RNA.
RNA editing:
- The coding sequence of an mRNA molecule is altered after transcription in RNA editing, so that the protein has an amino acid sequence that varies from that of the gene encoded.
- There were substitutions in some of the mRNA nucleotides in some nuclear genes in mammalian cells and in some mitochondrial genes in plant cells.
- More extensive RNA editing for certain mitochondrial genes in trypanosome parasites has been found in the mRNA.
- More than 60 percent of the sequence is determined by RNA editing in some of these organisms’ mRNAs.
- In RNA sequences, a variety of mechanisms can bring about changes.
- Molecules called guide RNAs (gRNAs) play a key role in certain situations.
- gRNAs consist of sequences that are partly complementary to pre-edited RNA segments.
- In these sequences, the two molecules goes through base pairing.
- The mRNA undergoes cleavage after the mRNA is anchored to the gRNA and nucleotides are inserted, removed or altered according to the gRNA template given.
- Enzymes bring about the conversion of the base in other cases.
- For example, in humans, a gene is transcribed into mRNA that encodes a lipid-transporting polypeptide called apolipoprotein-B100, which is synthesized in liver cells and has 4563 amino acids.
- By editing the apolipoprotein-B100 mRNA, a truncated version of the protein called apolipoprotein-B48 with only 2153 amino acids is synthesized in intestinal cells.
- A cytosine base is deaminated by an enzyme in this editing, transforming it into uracil.
- This conversion converts a codon that specifies the glutamine amino acid into a stop codon that terminates translation prematurely, resulting in the protein being shortened.
