
Translation in Prokaryotes
- It is the process of synthesis of protein by encoding information on mRNA.
- Protein synthesis requires mRNA, tRNA, aminoacids, ribosome and enzyme aminoacyl tRNA synthase
Various protein factors involved in protein synthesis
| Factors | Translation steps | Functions |
| IF-1 | Initiation | Helps to stabilize 30S ribosomal subunit |
| IF-2 | Initiation | Binds fmet-tRNA with 30S subunit mRNA complex; bind GTP and hydrolyse |
| IF-3 | Initiation | Binds 30S subunit with mRNA |
| EF-TU | Elongation | Binds GTP; bring Aminoacyl-tRNA to A-site of ribosome |
| EF-TS | Elongation | Generates EF-TU |
| EF-G | Elongation | Helps in translocation of ribosome |
| RF-1 | Termination | Helps to dissociates polypeptide from tRNA ribosome complex; specific for UAA and UAG |
| RF-2 | Termination | Helps to dissociates polypeptide; specific for UGA and UAA |
| RF-3 | Termination | Stimulates RF-1 and RF-2 |
Steps in translation:
1. Activation of aminoacids: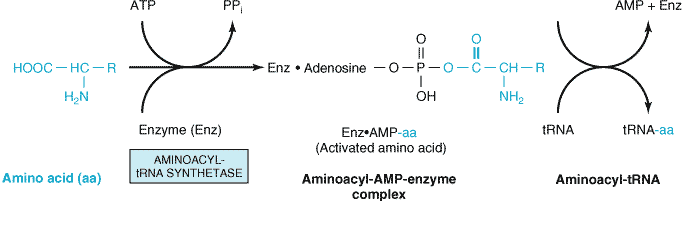
- The activation of aminoacids take place in cytosol.
- The activation of aminoacids is catalyzed by their aminoacyl tRNA synthetases.
- All the 20 aminoacids are activated and bound to 3’ end of their specific tRNA in the presence of ATP and Mg++.
- The N-formylated methionine is chain initiating aminoacid in bacteria whereas methionine is chain initiating aminoacid in eukaryotes.
- Methionine is activated by methionyl-tRNA synthetase. For N-formylmethionine two types of tRNA are used ie. tRNAmet and tRNAfmet.
- Similarly, all 2o aminoacids are activated (amino acyl-AMP enzyme complex) and then bound to their specific tRNA forming Aminoacyl tRNA.
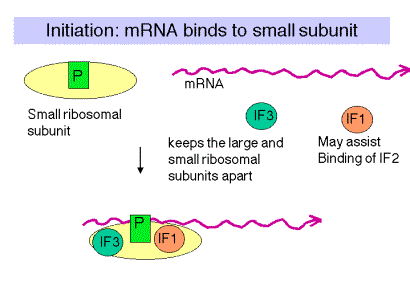
2. Initiation:
- In the first step, initiation factor-3 (IF-3) binds to 30S ribosomal unit.
- Then mRNA binds to 30S ribosomal subunit in such a way that AUG codon lie on the peptidyl (P) site and the second codon lies on aminoacyl (A) site.
- The tRNA carrying formylated methionine ie. FMet–tRNAFMet is palced at P-site. This specificity is induced by IF-2 with utilization of GTP. The IF-1 prevent binding of FMet–tRNAFMet is in A-site.
- Shinedalgrno sequence in the mRNA guide correct positioning of AUG codon at P-site of 30S ribosome.
- After binding of FMet–tRNAFMet on P-site, IF-3, IF-2 and IF-1 are released so that 50S ribosomal unit bind with 30S forming 70S sibosome. The exit site is located in 50S.
3. Elongation: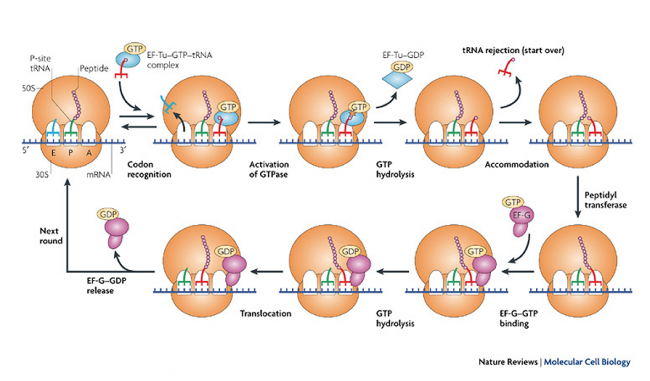
i. Binding of AA-tRNA at A-site:
- The 2nd tRNA carrying next aminoacid comes into A-site and recognizes the codon on mRNA. This binding is facilitated by EF-TU and utilizes GTP.
- After binding, GTP is hydrolysed and EF-TU-GDP is releasd
- EF=TU-GDP then and enter into EF-TS cycle.
ii. Peptide bond formation:
- The aminoacid present in t-RNA of P-site ie Fmet is transferred to t-RNA of A-site forming peptide bond. This reaction is catalyzed by peptidyltransferase.
- Now, the t-RNA at P-site become uncharged
iii. Ribosome translocation:
- After peptide bond formation ribosome moves one codon ahead along 5’-3’ direction on mRNA, so that dipeptide-tRNA appear on P-site and next codon appear on A-site.
- The uncharged tRNA exit from ribosome and enter to cytosol.
- The ribosomal translocation requires EF-G-GTP (translocase enzyme) which change the 3D structure of ribosome and catalyze 5’-3’ movement.
- The codon on A-site is now recognized by other aminoacyl-tRNA as in previous.
- The dipeptide on P-site is transferred to A-site forming tripeptide.
- This process continues giving long polypeptide chain of aminoacids.
4. Termination: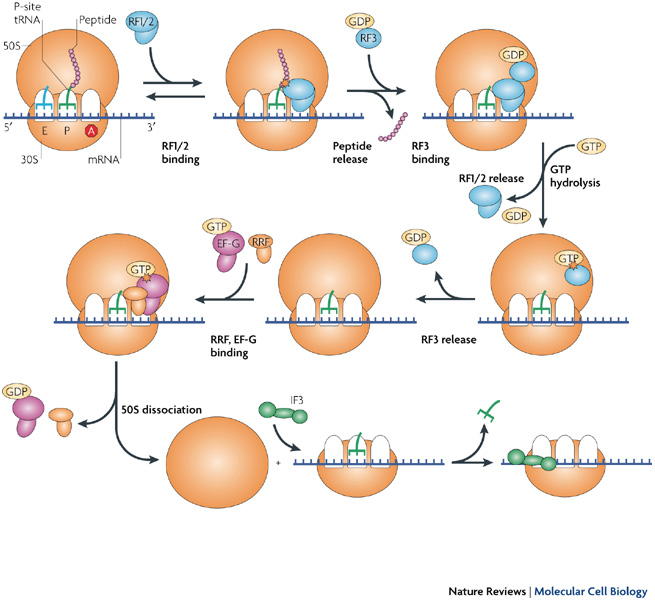
- The peptide bond formation and elongation of polypeptide continues until stop codon appear on A-site.
- If stop codon appear on A-site it is not recognized by t-RNA carrying aminoacids because stop codon donot have anticodon on mRNA.
- The stop codon are recognized by next protein called release factor (Rf-1, RF-2 and RF-3) which hydrolyses and cause release of all component ie 30s, 50S, mRNA and polypeptide separates.
- RF-1 recognisaes UAA and UAg while RF-2 recognises UAA and UGA while RF-3 dissociate 30S and 50S subunits.
- In case of eukaryotes only one release actor eRF causes dissociation.
Post translation modification:
The newly formed polypeptide may not be biologiy functional so it undergoes several folding and processing known as post translation modification.
1. Amino terminal and carboxyl terminal modification:
- The N-formylmethionine in case of bacteria is removed from polypeptide chain and some carboxyl terminal are also removed by enzymatic action to make functional protein. In case of eukaryotic protein, amino terminal is N- acetylated.
2. Loss of signal sequences:
- In some protein the amino terminal end is cleaved by specific peptidase so that protein loss its signaling property.
3. Modification of individual aminoacids:
- The aminoacids may be phosphorylated, acetylated for modification
4. Attachment of carbohydrate side chain:
- Carbohydrate side chain is added to make protein functional. Eg, glycoprotein. Lipoprotein
5. Addition of isoprenyl group:
- In some protein, isoprenyl group is added so to make protein active.
References
- https://courses.lumenlearning.com/microbiology/chapter/protein-synthesis-translation/
- https://www.khanacademy.org/science/biology/gene-expression-central-dogma/translation-polypeptides/a/the-stages-of-translation
- https://en.wikipedia.org/wiki/Prokaryotic_translation
- http://vle.du.ac.in/mod/book/print.php?id=13622&chapterid=30273
- http://www.biologydiscussion.com/cell/prokaryotes/translation-in-prokaryotes-genetics/38022
- http://www.sparknotes.com/biology/molecular/translation/section3.rhtml

Really informative and lucidly written