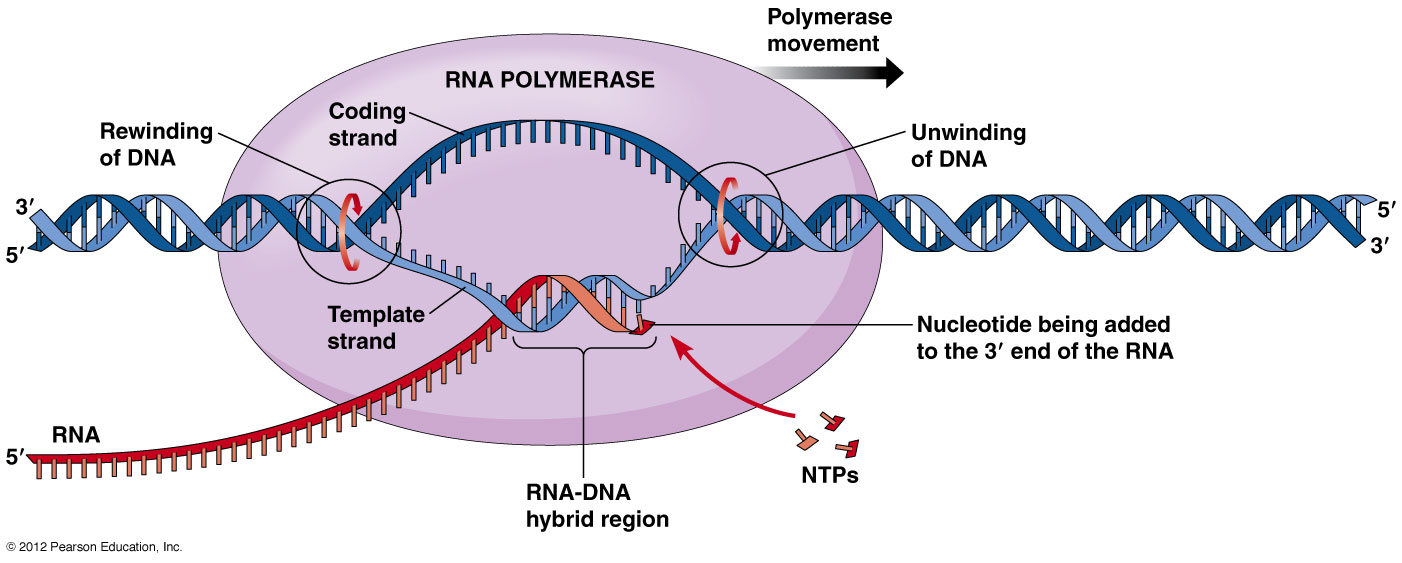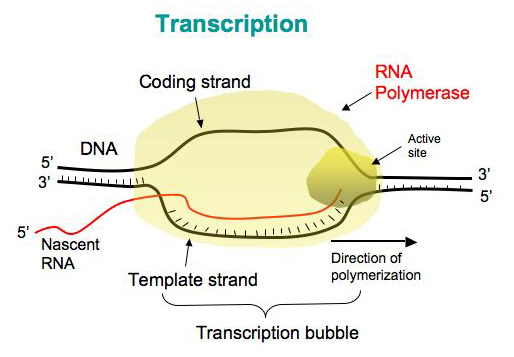
Transcription in Prokaryotes
- The process of synthesis of RNA by copying the template strand of DNA is called transcription.
- During replication entire genome is copied but in transcription only the selected portion of genome is copied.
- The enzyme involved in transcription is RNA polymerase. Unlike DNA polymerase it can initiate transcription by itself, it does not require primase. More exactly it is a DNA dependent RNA polymerase.
The steps of transcription
transcription is an enzymatic process. the mechanism of transcription completes in three major steps
1. Initiation:
- closed complex formation
- Open complex fromation
- Tertiary complex formation
2. Elongation
3. Termination:
- Rho- dependent
- Rho-independent
1. Initiation:
- The transcription is initiated by RNA polymerase holoenzyme from a specific point called promotor sequence.
- Bacterial RNA polymerase is the principle enzyme involved in transcription.
- Single RNA polymerase is found in a bacteria which is called core polymerase and it consists of α, β, β’ and ω sub units.
- The core enzyme bind to specific sequence on template DNA strand called promotor. The binding of core polymerase to promotor is facilitates and specified by sigma (σ) factor. (σ70 in case of E. coli).
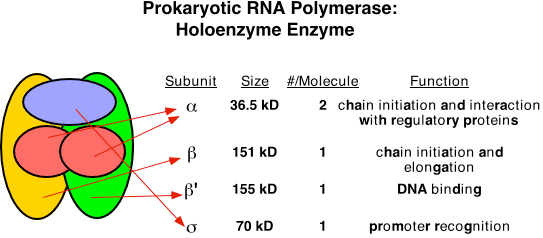
- The core polymerase along with σ-factor is called Holo-enzyme ie. RNA polymerase holoenzyme.
- In case of e. coli, promotor consists of two conserved sequences 5’-TTGACA-3’ at -35 element and 5’-TATAAT-3’ at -10 element. These sequence are upstream to the site from which transcription begins. Binding of holoenzyme to two conserve sequence of promotor form close complex.
- In some bacteria, the altered promotor may exist which contain UP-element and some may contain extended -10 element rather than -35 element.
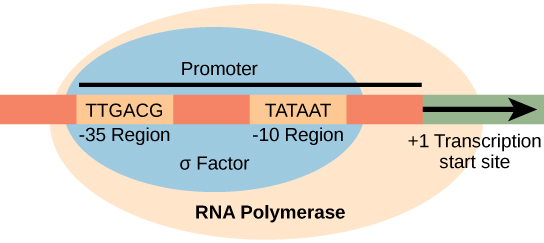
σ70 of E. coli has four region.
- Region1: it includes 1.2 and 1.1 region. Region 1.1 acts as molecular mimic of DNA
- Region2: it recognizes -10 element in promotor. α-helix recognizes -10 element.
- Region 3: it recognizes extended -10 element.
- Region 4: it recognizes -35 element in promotor by a structure called helix-turn-helix.
The UP-element is recognized by a carboxyl terminal domain of α-sub unit called αCTD (carboxyl terminal domain) which is connected to αNTD (Amino terminal domain) by flexible linker.
i. closed complex:
- Binding of RNA polymerase holoenzyme to the promotor sequence form closed comolex
ii. Open complex:
- after formation of closed complex, the RNA polymerase holoenzyme separates 10-14 bases exztending from -11 to +3 called melting. So that open complex is formed. This changing from closed complex to open complex is called isomerization.
iii. Tertiary complex:
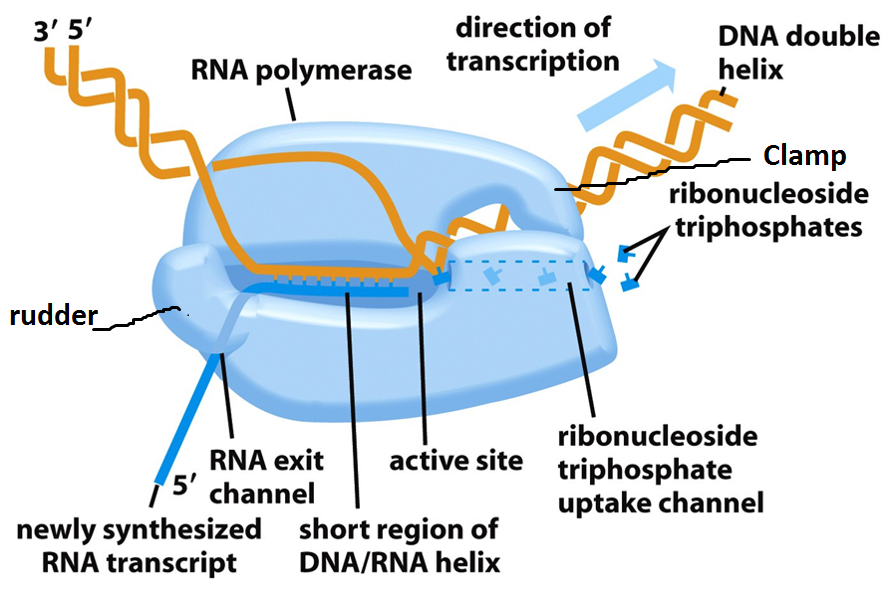
- RNA polymerase starts synthesizing nucleotide. It doesnot require the help of primase.
- If the enzyme synthesize short RNA molecules of less than 10 bp, it does not further elongates which is called abortive initiation. This is because σ 3.2 acting as mimic of RNA and it lies at middle of RNA exit channel in open complex.
- When the RNA polymerase manage to synthesize RNA more than 10 bp long, it eject the σ 3.2 region and RNA further elongates and exit from RNA exit channel. This is the formation of tertiary complex.
2. Elongation:
- After synthesis of RNA more than 10 bp long, the σ-factor is ejected and the enzyme move along 5’-3’ direction continuously synthesizing RNA.
- The synthesized RNA exit from RNA exit channel.
- The synthesized RNA is proof reads by Hydrolytic editing. For this the polymerase back track by one or more nucleotide and cleave the RNA removing the error and synthesize the correct one. The Gre factor enhance this proof reading process.
- Pyrophospholytic editing another mechanism of removing altered nucleotide.
3. Termination:
There are two mechanism of termination.
i. Rho independent: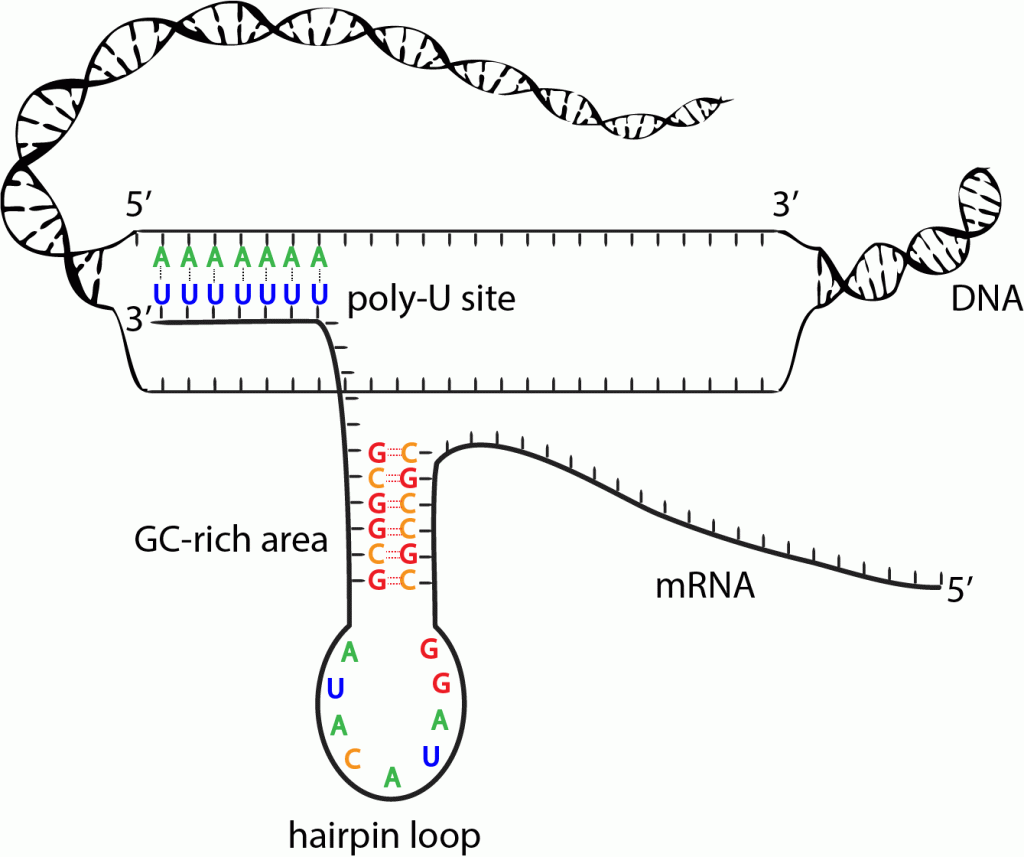
- In this mechanism, transcription is terminated due to specific sequence in terminator DNA.
- The terminator DNA contains invert repeat which cause complimentary pairing as transcript RNA form hair pin structure.
- This invert repeat is followed by larger number of TTTTTTTT(~8 bp) on template DNA. The uracil appear in RNA. The load of hair pin structure is not tolerated by A=U base pair so the RNA get separated from RNA-DNA heteroduplex.
ii. Rho dependent: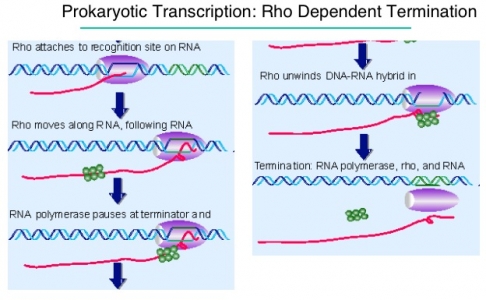
- In this mechanism, transcription is terminated by rho (ρ) protein.
- It is ring shaped single strand binding ATpase protein.
- The rho protein bind the single stranded RNA as it exit from polymerase enzyme complex and hydrolyse the RNA from enzyme complex.
- The rho protein does not bind to those RNA whose protein is being translated. Rather it bind to RNA after translation.
- In bacteria transcription and translation occur simultaneously so the rho protein bind the RNA after translation has completed but transcription is still ON.
References
- https://www.britannica.com/science/transcription-genetics
- https://en.wikipedia.org/wiki/Transcription_(biology)
- https://www.khanacademy.org/science/biology/gene-expression-central-dogma/transcription-of-dna-into-rna/a/overview-of-transcription
- https://www.nature.com/scitable/topicpage/dna-transcription-426
- https://www.ndsu.edu/pubweb/~mcclean/plsc731/transcript/transcript1.htm
- http://www2.le.ac.uk/projects/vgec/schoolscolleges/topics/geneexpression-regulation
- https://www.ck12.org/biology/Transcription-of-DNA-to-RNA/lesson/Transcription-of-DNA-to-RNA-BIO/
- http://oge.med.ufl.edu/courses/gms%206001/Transcription%20Prokaryotes%202012-c.pdf
- https://www.boundless.com/biology/textbooks/boundless-biology-textbook/genes-and-proteins-15/prokaryotic-transcription-107/initiation-of-transcription-in-prokaryotes-443-11667/
- https://www.ncbi.nlm.nih.gov/books/NBK9850/