Sanger’s method of gene sequencing
- Sanger’s method of gene sequencing is also known as dideoxy chain termination method. It generates nested set of labelled fragments from a template strand of DNA to be sequenced by replicating that template strand and interrupting the replication process at one of the four bases.
- Four different reaction mixtures are produced that terminates in A. T. G or C respectively.
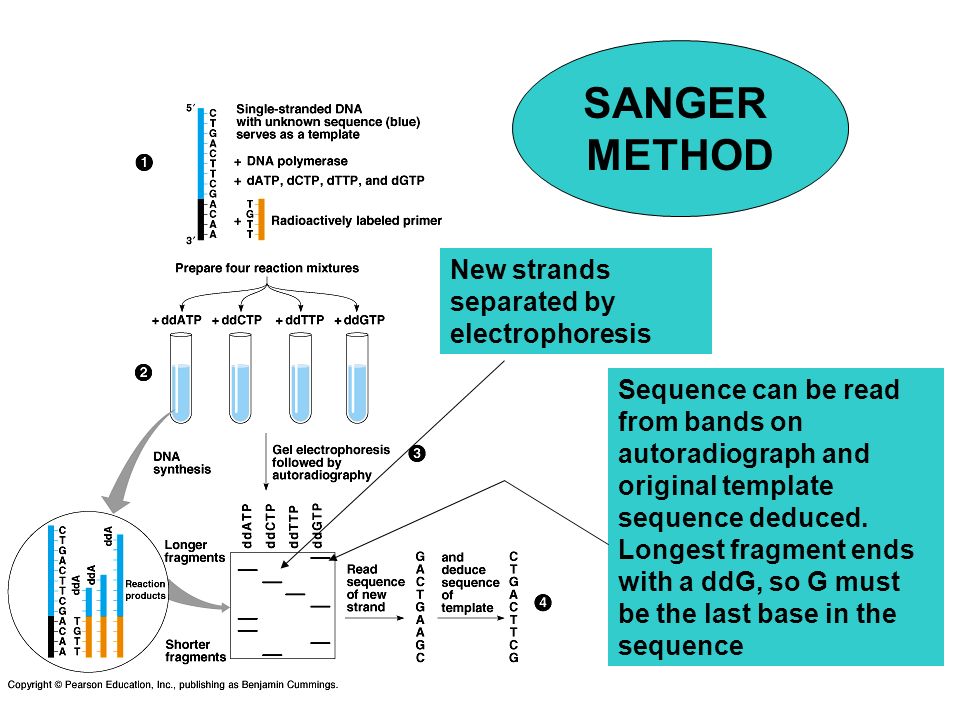
Principle
- A DNA primer is attached by hybridization to the template strand and deoxynucleosides triphosphates (dNTPPs) are sequentially added to the primer strand by DNA polymerase.
- The primer is designed for the known sequences at 3’ end of the template strand.
- M13 sequences is generally attached to 3’ end and the primer of this M13 is made.
- The reaction mixture also contains dideoxynucleoside triphosphate (ddNTPs) along with usual dNTPs.
- If during replication ddNTPs is incorporated instead of usual dNTPs in thegrowing DNA strand then the replication stops at that nucleotide.
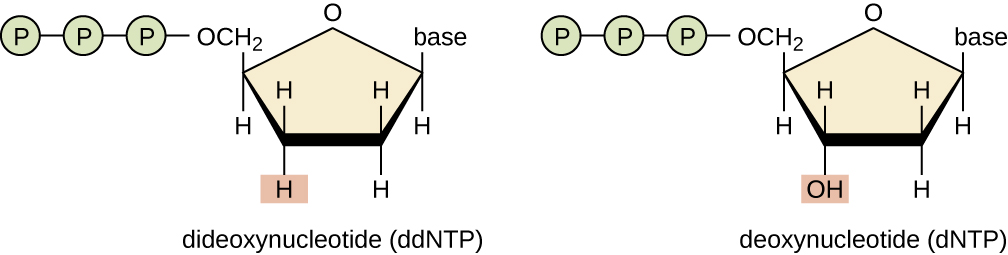
- The ddNTPs are analogue of dNTPs
- ddNTPs lacks hydroxyl group (-OH) at c3 of ribose sugar, so it cannot make phosphodiester bond with nest nucleotide, thus terminates the nucleotide chain
- Respective ddNTPs of dNTPs terminates chain at their respective site. For example ddATP terminates at A site. Similarly ddCTP, ddGTP and ddTTP terminates at C, G and T site respectively.
Procedure
1. Template preparation:
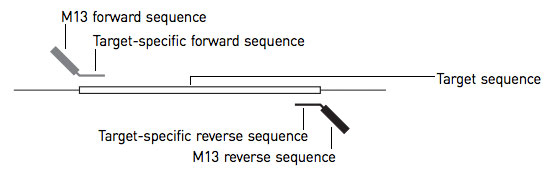
- Copies of template strand to be sequenced must be prepared with short known sequences at 3’ end of the template strand.
- A DNA primere is essential to initiate replication of template , so primer preparation of known sequences at 3’end is always required.
- For this purpose a single stranded cloning vector M13 is flanked with template strand at 3’end which serves as binding site for primer.
2. Generation of nested set of labelled fragments:
- Copies of each template is divided into four batches and each batch is used for different replication reaction.
- Copies of standard primer and DNA polymerase I are used in all four batches.
- To synthesize fragments that terminates at A, ddATP is added to the reaction mixture on batch I along with dATP, dTTP,dCTP and dGTP, standard primer and DNA polymerase I.
- Similarly, to generate, all fragments that terminates at C, G and T, the respective ddNTPs ie ddCTP,ddGTP and ddTTP are added respectively to different reaction mixture on different batch along with usual dNTPs.
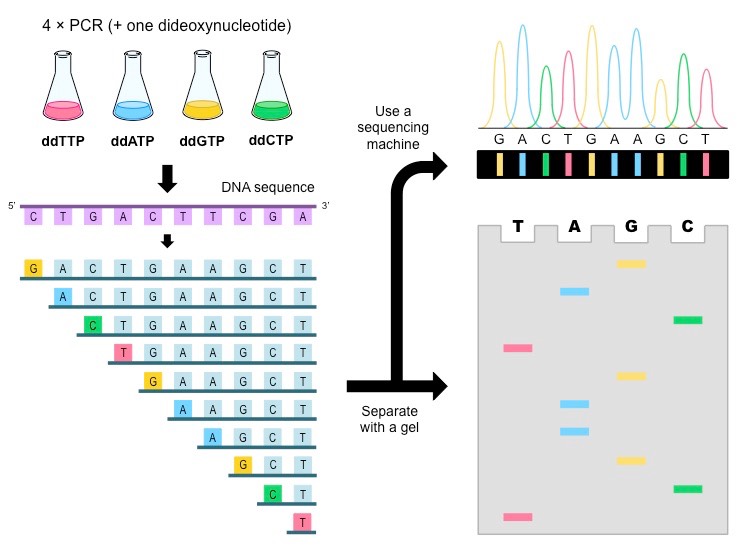
3. Electrophoresis and gel reading:
- The reaction mixture from four batches are loaded into four different well on polyacrylamide gel and electrophoresed.
- The autoradiogramof the gel is read to determine the order of bases of complementary strand to that of template strand.
- The band of shortest fragments are at the bottom of autoradiogram so that the sequences of complementary strand is read from bottom to top.
References
- https://www.boundless.com/microbiology/textbooks/boundless-microbiology-textbook/microbial-genetics-7/bioinformatics-83/dna-sequencing-based-on-sanger-dideoxynucleotides-459-6637/
- http://www.hhmi.org/biointeractive/sanger-method-dna-sequencing
- https://www.dnalc.org/view/15479-Sanger-method-of-DNA-sequencing-3D-animation-with-narration.html
- https://www.khanacademy.org/science/biology/biotech-dna-technology/dna-sequencing-pcr-electrophoresis/a/dna-sequencing
- https://www.thermofisher.com/np/en/home/life-science/sequencing/sanger-sequencing/sanger_sequencing_method.html
- https://en.wikipedia.org/wiki/Sanger_sequencing
- http://www.ithanet.eu/ithapedia/index.php/Protocol:DNA_sequencing
